标签:DNA sequence int pos depth 长度 hdu1560DNA IDA
题目描述:
DNA sequence
Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Total Submission(s): 6788 Accepted Submission(s): 3052
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
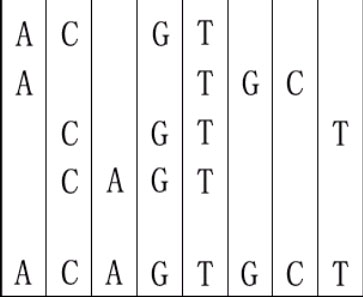
Input The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
Output For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
Sample Input 1 4 ACGT ATGC CGTT CAGT
Sample Output 8 题目大意:有许多序列,要把他们横着排列起来,相同的元素可以排在一行,求最终排列的最少长度 思路:咱们最终排列的长度不知道多长,然后排列的先后顺序也不是很清楚,如果单纯dfs,很可能超时/爆栈, 这时候就需要用到高级的玩意儿,迭代加深搜索,我们可以预设一个搜索的排列长度,我们就再这个长度下 遍历所有情况,如果都不满足就继续加大深度,不怕爆栈而且也不会一条路走到黑,需要注意的是那个用来回溯 的数组一定要再dfs里面定义(别问我怎么知道的...),具体可以看代码注释 AC代码:
#include<bits/stdc++.h>
using namespace std;
typedef long long ll;
const int maxn = 100;
const int inf = 0x3f3f3f3f;
string c = "ACGT";
string a[maxn];
int pos[10],n;
int getlast() {//所有字符串还没有放置长度的最大值,即剩余需要的step的最小可能值
int ans = 0;
for (int i = 0; i < n; i++) {
ans = max(ans, int(a[i].size()) - pos[i]);
}
return ans;
}
int depth;
bool dfs(int step) {
int last = getlast();
if (step + last>depth)return false;//剩余需要放的长度+已放置了的长度>预设的depth,A*剪枝
if (!last)return true;//如果没有剩余的需要放了,满足要求
int tmp[10];//别当成全局变量了...
memcpy(tmp, pos, sizeof(pos));
for (int i = 0; i < 4; i++) {//该层放什么元素
bool f = 0;
for (int j = 0; j < n; j++) {
if (pos[j]<int(a[j].size())&&a[j][pos[j]] == c[i]) {
//如果该字符串下一个放置的字符就是当前遍历的字符,就可以放置
f = 1;
pos[j]++;
}
}
if (f) {//如果该层可以放置
if (dfs(step + 1))return true;
memcpy(pos, tmp, sizeof(tmp));//回溯
}
}
return false;
}
int main() {
//freopen("test.txt", "r", stdin);
int t; scanf("%d", &t);
while (t--) {
scanf("%d", &n);
int maxlen = 0;
for (int i = 0; i < n; i++) {
cin >> a[i]; maxlen = max(maxlen, int(a[i].size()));
pos[i] = 0;
}
depth = maxlen;//预设初始深度为最长的字符串
while (1) {
if (dfs(0)) break;
depth++;
}
printf("%d\n", depth);
}
return 0;
}
/*
1
4
ACGT
ATGC
CGTT
CAGT
*/
/*
8
*/
标签:DNA,sequence,int,pos,depth,长度,hdu1560DNA,IDA 来源: https://www.cnblogs.com/MYMYACMer/p/14594815.html
本站声明: 1. iCode9 技术分享网(下文简称本站)提供的所有内容,仅供技术学习、探讨和分享; 2. 关于本站的所有留言、评论、转载及引用,纯属内容发起人的个人观点,与本站观点和立场无关; 3. 关于本站的所有言论和文字,纯属内容发起人的个人观点,与本站观点和立场无关; 4. 本站文章均是网友提供,不完全保证技术分享内容的完整性、准确性、时效性、风险性和版权归属;如您发现该文章侵犯了您的权益,可联系我们第一时间进行删除; 5. 本站为非盈利性的个人网站,所有内容不会用来进行牟利,也不会利用任何形式的广告来间接获益,纯粹是为了广大技术爱好者提供技术内容和技术思想的分享性交流网站。
